Omics
Genomics Technology Enables Quick, Cost-Effective Sampling for Better Results
SCROLL TO LEARN MORE
What We Do
AOML scientists use Omics as an umbrella term for the study of various fields such as genomics, metagenomics, metatranscriptomics, proteomics, metabolomics, epigenomics, and high-throughput amplicon sequencing. These emerging fields help us answer research questions about DNA, RNA, proteins, and other small molecules from organisms and the environment. With this information, we can identify mechanisms that keep marine systems healthy and productive.
The Omics program at AOML works to promote coral resilience, develop and transfer emerging technologies, advance Omics for fisheries and microbiome applications, and foster the bioinformatics and infrastructure capabilities upon which all Omics research and operations rely. This work engages coral, fisheries, and microbiology experts across the agency and through international engagement.
The Five Pillars of AOML’s Omics Activities
Areas of Research
Who We Are
Coral Restoration Omics
Resilience and Resistance
Our research aims to identify what makes certain corals more resilient to stressors like heat, ocean acidification and disease. Understanding the molecular underpinnings of coral resilience and susceptibility by identifying resilient genotypes allows resource managers to be more effective with their restoration plans, especially when out-planting corals on the reef.
Heat Stress Resilience
AOML is working to identify resilient coral genotypes to isolate the molecular mechanisms by which resilience is transferred through colonies. In-depth field research of coral bleaching patterns has revealed that some corals are more resistant to bleaching from heat stress.
Those corals have particular genetic signatures that are have acclimated to +1.0°C hotter than others of the same species. Researchers may be able to use this information to help build more resilient reefs for the future.
By identifying genetic markers for coral resilience both through lab research and field research, we can understand bleaching responses and be proactive when environmental stressors affect our reef ecosystems.
Resistance to Disease
Part of our research addresses the devastating consequences of disease spreading through South Florida and the Florida Keys at the molecular level. Scientists are modeling how disease spreads through a reef to improve their prediction of the spread of disease.
With this information, researchers can help to improve resistance on the reef and evaluate the longevity of different types of corals on the reef. Microbial source tracking is also being used to track land-based sources of pollution, and DNA sequencing is being used to investigate the biodiversity of coral microbiomes.
Advancing eDNA
eDNA Can Aid in the Detection of Higher Trophic Levels
eDNA Can Inform Our Understanding of the Microbiome
eDNA Is Next Generation Sustainable Science
To Understand the Microbiome
There is a global need for low-cost indicators that measure the health of marine ecosystems. The microbiome forms the base of the food web and controls things like carbon, nutrients, metals and toxins – that means that we can’t truly understand the state of marine ecosystems until we have a good understanding of its microbiome. This understanding has allowed us to appreciate the role of microbes in all aspects of life, including ocean resiliency and marine resource management as well as and infection and disease.
To Detect Higher Trophic Levels
Because eDNA comes from cells that have been sloughed or excreted from a marine organism, eDNA offers a unique opportunity to detect what marine organisms have been in the area using only seawater samples. This can be especially useful in remote sensitive areas. It also offers the ability to detect multiple trophic levels from a single sample.
We are working on using environmental DNA (eDNA) – DNA from filtered seawater to understand what kinds of higher trophic level animals (like fish or mammals) are found in a particular area. This is done by capture of sloughed or excreted cells. eDNA is collected from seawater or sediment instead of from tissues so that samples can be collected without animal capture, tissue processing, or trawling through sensitive habitats.
Exploring Environmental DNA Video Series
Research involving Environmental DNA or “eDNA” is an exciting and emerging area of science that can help scientists to manage endangered species, invasive species, and monitor the biodiversity of ecosystems. Learn about environmental DNA through the “Exploring Environmental DNA” video series that covers what “eDNA” is, environmental DNA sampling technology developed at NOAA AOML, and a hands-on activity for DNA extraction.
Omics for Fisheries & Protected Species
eDNA for Protected Marine Turtles
Meeting Monitoring Requirements with Efficiency and Minimal Impact
A population of ~60 ‘black’ green turtles (Chelonia mydas agassizii) were listed as an endangered species and have been monitored since the 1970’s under the Endangered Species Act at the South San Diego Bay National Wildlife Refuge. Regular and continued monitoring of this population size is needed to support the National Marine Fisheries Service recovery plan for U.S. Pacific green turtle populations under shifting habitat use. Traditional monitoring methods are high-impact, laborious and expensive. By utilizing eDNA technology, low-impact spatial mapping may be possible.
Check out the web story published on NOAA Research.

Fisheries Genomics
Our ability to effectively manage fisheries is limited by our understanding of fishery populations and their dependence on environmental conditions. Including genomic information into fisheries management may improve the decision-making process and thus the sustainability of fisheries.
How genomic information affects decisions will be evaluated using advanced population assessment techniques, and the Management Strategy Evaluation framework adopted by the National Marine Fisheries Service. AOML and the Southeast Fisheries Science Center (SEFSC) have formed a collaborative partnership with a clear path to transition results into fisheries management plans for the following projects:
- Using DNA Single Nucleotide Polymorphisms (DNA SNPs) to differentiate between eastern and western stocks of Atlantic bluefin tuna to inform stock assessments and management strategy evaluation.
- Using Restriction site Associated DNA Sequencing (ddRAD-Seq) to access the population structure of king mackerel to address pressing uncertainties affecting the stock assessment of this species.
- Empirical models are being used to assess whether microbiome data (sometimes termed “eDNA”) improves the ability to predict distributions of fish and to understand trophic interactions and habitat use.
Enhancing Omics Workforce & Infrastructure
Experimental Reef Laboratory
The Experimental Reef Laboratory located on the University of Miami’s Virginia Key campus at the Rosenstiel School of Marine and Atmospheric Sciences was completed in September of 2016. This unique experimental facility was designed to study the combined effect of heat stress and ocean acidification on corals so that scientists can see how coral organisms respond at the molecular level (DNA and RNA) under present and possible future conditions.
Rollover the dots to learn more or visit Experimental Reef Lab’s Page.
By building in-house facilities such as these, we can fulfill critical gaps in in equipment, increase efficiency and integrity of sample processing, and offer a place for partners (such as the Southeast Fisheries Science Center) and others to collaborate on new research projects.
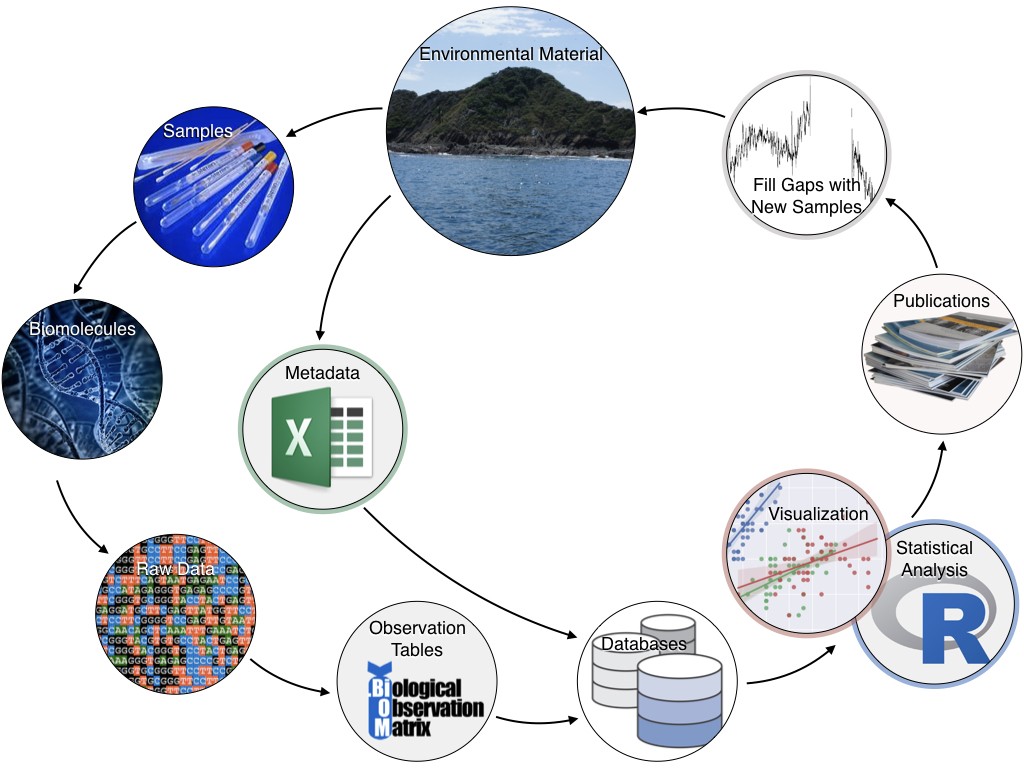
Interpreting Omics Data with Bioinformatics
Genome-based techniques improve our ability to characterize and monitor ecosystems, including characterization environments with potential commercial potential. Examples include genomic signatures that mark natural resources such as oil and gas reserves, enzymes with commercial potential such as pharmaceutical or bioremediation potential, microbiomes that control the health of commercially valuable wild and aquaculture species, and indicators of anthropogenic stress which can forecast degraded environmental productivity.
However, our ability to supply bioinformatics expertise has not kept pace with the generation of sequence data, creating a data backlog and hindering transition of data collected into actionable information. To address this gap, AOML has been working to develop bioinformatics capacity, which is important to the success of all ‘omics projects. AOML has secured servers dedicated to bioinformatic analysis, hired young scientists to help with analysis, and created user groups (local and NOAA-wide) to provide support.
To facilitate analysis of amplicon datasets, a bioinformatics pipeline (“Tourmaline”) was created and added to AOML’s GitHub account. This automated amplicon workflow uses QIIME 2 and Snakemake and can run Deblur (single-end) or DADA2 (single-end and paired-end); steps can be manually adjusted if needed.
Publications & References
Thompson, L.R., M.F. Haroon, A.A. Shibl, M.J. Cahill, D.K. Ngugi, G.J. Williams, J.T. Morton, R. Knight, K.D. Goodwin & U. Stingl. (2019). Red Sea SAR11 and Prochlorococcus single-cell genomes reflect globally distributed pangenomes. Appl Environ Microbiol, https://doi.org/10.1128/AEM.00369-19.
Franzosa, E.A., L.J. McIver, G. Rahnavard, L.R. Thompson, M. Schirmer, G. Weingart, K. Schwarzberg Lipson, R. Knight, J.G. Caporaso, N. Segata & C. Huttenhower. (2018). Species-level functional profiling of metagenomes and metatranscriptomes. Nat Meth, https://doi.org/10.1038/s41592-018-0176-y.Thompson, L.R., J.G. Sanders, D. McDonald, A. Amir, J. Ladau, K.J. Locey, R.J. Prill, A. Tripathi, S.M. Gibbons, G.Ackermann, J.A. Navas-Molina, S. Janssen, E. Kopylova, Y. Va ́zquez-Baeza, A. Gonza ́lez, J.T. Morton, S. Mirarab, Z.Z. Xu, L. Jiang, M.F. Haroon, J. Kanbar, Q. Zhu, S. Song, T. Kosciolek, N.A. Bokulich, J. Lefler, C.J. Brislawn, G.C. Humphrey, S.M. Owens, J. Hampton-Marcell, D. Berg-Lyons, V. McKenzie, N. Fierer, J.A. Fuhrman, A. Clauset, R.L. Stevens, A. Shade, K.S. Pollard, K.D. Goodwin, J.K. Jansson, J.A. Gilbert, R. Knight & The Earth Microbiome Project Consortium. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463, https://doi.org/10.1038/nature24621.Goodwin, K.D., L.R. Thompson, B. Duarte, T. Kahlke, J.C. Marques & I. Cac ̧ador. (2017). DNA sequencing as atool to monitor marine ecological status. Front Mar Sci 4:107, https://doi.org/10.3389/fmars.2017.00107.Amir, A., D. McDonald, J.A. Navas-Molina, E. Kopylova, J. Morton, Z.Z. Xu, E.P. Kightley, L.R. Thompson, E.R. Hyde, A. Gonzalez & R. Knight. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2(2):e00191–16, https://doi.org/10.1128/mSystems.00191-16.
Looking for Literature? Search our Publication Database.





















