Dr. Nastassia Patin, a University of Miami Cooperative Institute for Marine and Atmospheric Studies (CIMAS) scientist working at AOML, recently spent three weeks aboard the NOAA ship Reuben Lasker collecting environmental DNA (eDNA) from water samples in support of the Rockfish Recruitment and Ecosystem Assessment Survey (RREAS).
Environmental DNA (eDNA) refers to DNA collected directly from habitats like soil, freshwater, or the ocean. It contains DNA from organisms that live in or have passed through that habitat, including rare, invasive, or endangered species.

In collaboration with scientists from the National Marine Fisheries Service, Dr. Patin collected samples corresponding to nighttime midwater trawls at transect stations along the coast of California, from Point Reyes to San Diego.
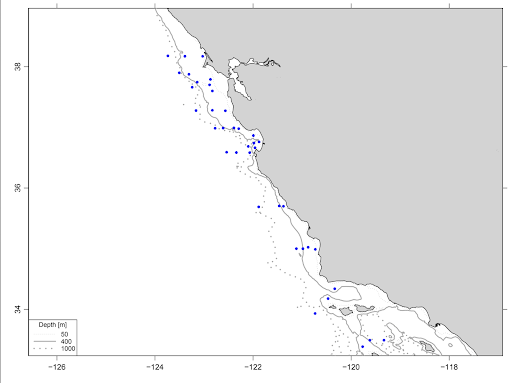
Dr. Patin collected a total of 339 water samples that were filtered and preserved for eDNA.
“At each station I collected triplicate water samples from three depths using a CTD and Niskin bottle rosette. I filtered each water sample through a 0.22 um membrane filter and froze the filters at -80°C,” explained Dr. Patin.
A CTD (conductivity, temperature, and depth) is an instrument used to detect changes in conductivity and water temperature relative to depth. A CTD is attached to a rosette (metal water sampling array) and Niskin bottles (for multiple water samples) that are lowered into the water with a cable. When the rosette is deployed, the open bottles are triggered to close and collect water samples at various depths.
The eDNA will be used for marker gene sequencing (“metabarcoding”) for organisms at different trophic levels from microbes to whales. Trophic level refers to the position of an organism in a food chain, based on the amount of consumption steps that separates it from the original energy source, such as the sun. Interactions between organisms in different trophic levels cause the energy flow to be more complex than a linear chain (food chain), and is better represented as a “food web.”
Additionally, eDNA will be analyzed to provide information on microbial genomes and biochemical function, which is not attained through metabarcoding.

“Marine eDNA research has lagged behind that of terrestrial and aquatic environments, largely due to challenges with field sampling and sequence data analysis,” said Dr. Patin.
As part of NOAA’s efforts to develop eDNA methods, Dr. Patin worked to optimize sampling protocols aboard the Reuben Lasker, a challenging project considering the ship was not designed for sensitive molecular work. Advancements in eDNA sampling and analysis methods will improve detection of rare, endangered, or invasive species that may not be comprehensively assessed with visual surveys.
“Expanding macrofaunal assessments beyond visual surveys will provide more precise information on areas of predator/prey overlap than is currently possible with visual surveys,” said Dr. Patin. “Further, linking multiple trophic levels will contribute to our understanding of coastal marine food webs, whose foundation is microbial and the highest levels of which include blue whales.”

Environmental DNA data generated from the RREAS survey will be sequenced and analyzed with trawl catch information and other ecological metrics to provide a better understanding of marine food webs, organism interconnectivity in the the California Current, and prey/predator coexistence in these areas.
